Department of Health and Human Services
National Institutes of Health
National Library of Medicine (NLM)
2019 Congressional Justification (PDF)
FY 2019 Budget
- Organization Chart
- Appropriation Language
- Amounts Available for Obligation
- Budget Graphs
- Authorizing Legislation
- Appropriations History
- Justification of Budget Request
- Detail of Full-Time Equivalent Employment (FTE)
- Detail of Positions
Organizational Chart

- Office of the Director
Patricia Flatley Brennan, R.N., Ph.D., Director
Jerry Sheehan, Deputy Director
Milton Corn, M.D., Deputy Director for Research and Evaluation
Todd D. Danielson, Associate Director for Administrative Management- Division of Extramural Programs
Valerie Florance, Ph.D., Director - Division of Library Operations
Joyce E. B. Backus, Associate Director - Lister Hill National Center for Biomedical Communications
Clem McDonald, M.D., Director - Division of Specialized Information Services
Florence Chang, Acting Associate Director - National Center for Biotechnology Information
James M. Ostell, Ph.D., Director
- Division of Extramural Programs
Appropriation Language
For carrying out section 301 and title IV of the PHS Act with respect to health information communications, $395,493,000: Provided, That of the amounts available for improvement of information systems, $4,000,000 shall be available until September 30, 2020: Provided further, That in fiscal year 2019, the National Library of Medicine may enter into personal services contracts for the provision of services in facilities owned, operated, or constructed under the jurisdiction of the National Institutes of Health (referred to in this title as “NIH”).
Amounts Available for Obligation 1
(Dollars in Thousands)
| Source of Funding | FY 2017 Final | FY 2018 Annualized CR | FY 2019 President's Budget |
|---|---|---|---|
| Appropriation | $407,510 | $407,510 | $395,493 |
| Mandatory Appropriation: (non-add) | |||
| Type 1 Diabetes | (0) | (0) | (0) |
| Other Mandatory financing | (0) | (0) | (0) |
| Rescission | 0 | -2,767 | 0 |
| Sequestration | 0 | 0 | 0 |
| Secretary's Transfer | -906 | ||
| Subtotal, adjusted appropriation | $406,604 | $404,743 | $395,493 |
| OAR HIV/AIDS Transfers | 0 | 0 | 0 |
| Subtotal, adjusted budget authority | $406,604 | $404,743 | $395,493 |
| Unobligated balance, start of year | 2,000 | 2,200 | 0 |
| Unobligated balance, end of year | -2,200 | 0 | 0 |
| Subtotal, adjusted budget authority | $406,404 | $406,943 | $395,493 |
| Unobligated balance lapsing | -2,354 | 0 | 0 |
| Total obligations | $404,050 | $406,943 | $395,493 |
1 Excludes the following amounts (in thousands) for reimbursable activities carried out by this account:
FY 2017 - $22,230 FY 2018 - $22,230 FY 2019 - $18,274
Fiscal Year 2019 Budget Graphs
History of Budget Authority and FTEs:
Bar Graph for Funding Levels by Fiscal Year for FY2015 through FY2019*
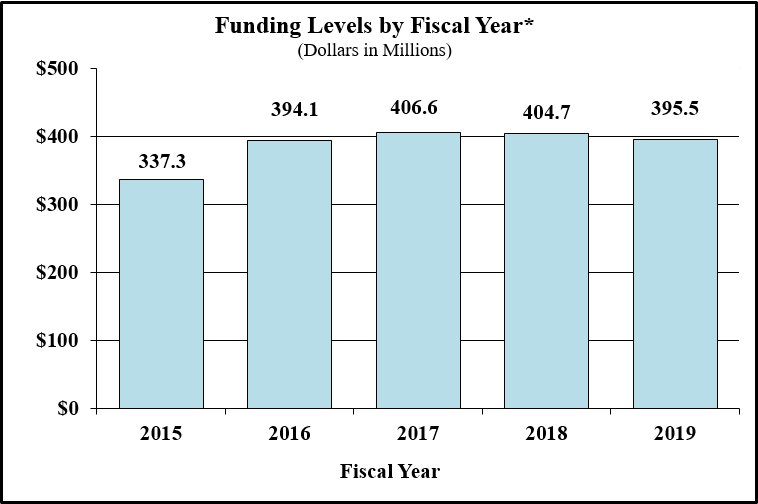
Data for Funding Levels by Fiscal Year for FY2015 through FY2019*
| Fiscal Year | Funding (Dollars in Millions) |
|---|---|
| 2015 | 337.3 |
| 2016 | 394.1 |
| 2017 | 406.6 |
| 2018CR | 404.7 |
| 2019PB | 395.5 |
Bar Graph of FTEs by Fiscal Year for FY2015 through FY2019*
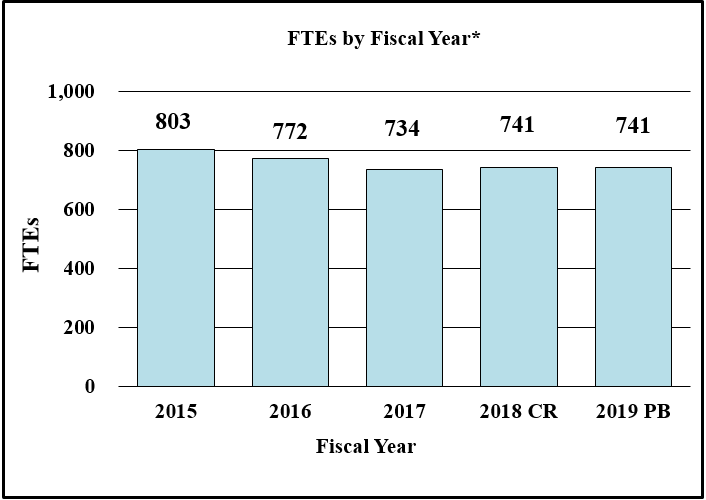
Data for FTEs by Fiscal Year for FY2015 through FY2019*
| Fiscal Year | FTEs |
|---|---|
| 2015 | 803 |
| 2016 | 772 |
| 2017 | 734 |
| 2018CR | 741 |
| 2019PB | 741 |
*Note: These numbers are formula driven.
Authorizing Legislation
| PHS Act/ Other Citation |
U.S. Code Citation |
2018 Amount Authorized |
FY 2018 Annualized CR | 2019 Amount Authorized |
FY 2019 President's Budget1 | |
|---|---|---|---|---|---|---|
| Research and Investigation | Section 301 | 42§241 | Indefinite | $404,742,600 | Indefinite | $395,493,000 |
| National Library of Medicine | Section 401(a) | 42§281 | Indefinite | Indefinite | ||
| Total, Budget Authority | $404,742,600 | $395,493,000 | ||||
Appropriations History
| Fiscal Year | Budget Estimate to Congress | House Allowance | Senate Allowance | Appropriation |
|---|---|---|---|---|
| 2009 | $323,046,000 | $331,847,000 | $329,996,000 | $330,771,000 |
| Rescission | $0 | |||
| Supplemental | $1,705,000 | |||
| 2010 | $334,347,000 | $342,585,000 | $336,417,000 | $339,716,000 |
| Rescission | $0 | |||
| 2011 | $364,802,000 | $364,254,000 | $339,716,000 | |
| Rescission | $2,982,909 | |||
| 2012 | $387,153,000 | $387,153,000 | $358,979,000 | $338,278,000 |
| Rescission | $639,345 | |||
| 2013 | $372,651,000 | $381,981,000 | $337,638,655 | |
| Rescission | $675,277 | |||
| Sequestration | ($16,947,139) | |||
| 2014 | $382,252,000 | $387,912,000 | $327,723,000 | |
| Rescission | $0 | |||
| 2015 | $372,851,000 | $336,939,000 | ||
| Rescission | $0 | |||
| 2016 | $394,090,000 | $341,119,000 | $402,251,000 | $394,664,000 |
| Rescission | $0 | |||
| 20171 | $395,684,000 | $407,086,000 | $412,097,000 | $407,510,000 |
| Rescission | $0 | |||
| 2018 | $373,258,000 | $413,848,000 | $420,898,000 | $407,510,000 |
| Rescission | $2,767,400 | |||
| 2019 | $395,493,000 |
1Budget Estimate to Congress includes mandatory financing.
Justification of Budget Request
Authorizing Legislation: Section 301 and title IV of the Public Health Service Act, as amended.
Budget Authority (BA):
| FY 2017 Actual |
FY 2018 Annualized CR |
FY 2019 President’s Budget |
FY 2019 +/- FY 2018 |
|
|---|---|---|---|---|
| BA | $406,604,000 | $404,742,600 | $395,493,000 | -$9,249,600 |
| FTE | 734 | 741 | 741 | 0 |
Program funds are allocated as follows: Competitive Grants/Cooperative Agreements; Contracts; Direct Federal/Intramural and Other.
Director's Overview
The National Library of Medicine (NLM), the world’s largest biomedical library, acquires, organizes, and delivers up-to-date biomedical information across the United States and around the globe. Millions of scientists, health professionals, and members of the public use NLM’s electronic information sources billions of times each year. Through its information systems, cutting-edge data science and informatics research, and extensive research training programs, NLM plays an essential role in catalyzing basic biomedical science. NLM makes research results available for translation into new treatments, products, and practices; provides useful decision support for health professionals and patients; and supports disaster and emergency preparedness and response. Leveraging its 180-year history of organizing and curating the biomedical literature, NLM is now expanding to other knowledge resources, particularly data, providing leadership in the acquisition and analysis of data for discovery, as well as training of biomedical data scientists.
Its strategic plan for 2017-2027 positions NLM to address existing and emerging challenges in biomedical research and public health. It will do this by expanding its core biomedical literature and genomic collections to include a broad array of health and biological data types and making these data findable, accessible, interoperable, and reusable. NLM will enhance its programs of research to systematically characterize and curate data reflecting complex health phenomena from cells to society and to devise new methodologies that uncover the knowledge held in data. NLM will expand its training programs to incorporate data science and maintain its commitment to outreach excellence and support of a diverse workforce. NLM will create an effective, efficient organizational structure for data science. NLM leadership re-affirms its commitment to act in concert with values across biomedical research, the NIH, libraries, and the public.
Priorities for FY 2019
Information Partnerships
The rich set of information resources provided by NLM are built on important partnerships with the private and public sectors. This includes partnerships with publishers of biomedical journals to submit data directly to PubMed and PubMed Central (PMC); partnerships in providing database content; partnerships in use of NLM services as platforms for compliance with policies such as public access policies; and partnerships in promoting data standards that enable data sharing. Partnerships are also important in the application of NLM resources to address public health priorities. For example, the pathogen detection project is a partnership with the Centers for Disease Control and Prevention (CDC), the Food and Drug Administration (FDA), the U.S. Department of Agriculture (USDA), Public Health England, and state and regional laboratories. NLM provides rapid analysis of genome sequences from field specimens to detect the most closely related sequences, enabling faster detection and response to outbreaks. Genome analytics are also used to optimize the recognition of antimicrobial resistance genes in genomic DNA in a partnership with FDA, CDC, and the Department of Defense (DOD).
Basic Research
NLM’s richly linked databases promote scientific breakthroughs by playing an essential role in all phases of research and innovation. Every day, NLM receives up to 12 Terabytes of new data and publications, adds value by enhancing quality and consistency and integrating them with other information in NLM databases, and responds to millions of inquiries from individuals and computer systems by serving up some 100 Terabytes of information, including 3 million published articles. New algorithms stemming from information science research have yielded enhancements in search methods to locate articles relevant to a search and related to each other.
Scientists around the world analyze data and information using software, algorithms, and methods arising from basic research in computational biology and biomedical informatics conducted or funded by NLM. For example, such tools are used to mine journal articles and electronic health records (EHRs) to discover adverse drug reactions, to analyze high throughput genomic data to identify promising drug targets, and to detect transplant rejection earlier so interventions to help clinical research participants can begin more quickly. Computational genomics research at NLM has led to insights for understanding a variety of issues, such as genetic mutational patterns and factors in disease, gene regulation processes, molecular binding, and protein structure and function. For example, a major breakthrough in gene editing technology, CRISPR-Cas, had a foundation in research on bacterial evolutionary genomics by intramural scientists at NLM.
Translational and Clinical Research to Improve Health
NLM supports both the discovery and direct clinical application of knowledge to improve health through its research, databases, data standards resources, and data analysis tools. The Database of Genotypes and Phenotypes (dbGaP) includes the results of more than 900 studies of the interaction between genetic makeup and observable traits (e.g., high cholesterol) associated with certain diseases. The ClinVar database makes knowledge about the clinical significance of scientifically validated genetic variations available to clinicians. ClinicalTrials.gov includes an increasing number of studies that identify genetic variations associated with different outcomes to provide confirming evidence for precision medicine. Biomedical informatics research funded and conducted by NLM is yielding advanced analytical methods and tools for use against large scale data generated from clinical care, leading to fuller understanding of the effects of medications and procedures as well as individual factors important in the prevention and treatment of disease processes.
A Diverse and Talented Research Workforce
NLM remains the primary federal funder of research training for biomedical informatics and data science. In support of the NIH Next Generation Researcher initiative, NLM places priority on research project awards to early stage investigators and early established investigators. To expand workforce capacity for advanced research in data science, NLM is supporting focused training and curriculum development in the area of biomedical data sciences. A lack of diversity exists in many research and scientific fields, including biomedical informatics and data science. In addition to outreach initiatives designed to engage members of underrepresented minorities in NLM’s university-based training programs, new incentives are being devised to recruit and retain underrepresented minorities, and also to increase the visibility and interest in biomedical informatics. NLM also hosts intramural postdoctoral and other professional-level training programs in biomedical informatics, data science, and bioinformatics.
Program Descriptions and Accomplishments
Intramural Programs
NLM’s intramural programs support both high-value information services and research that focuses on computational biology, biomedical informatics, data science, and information science. NLM is home to the National Center for Biotechnology Information (NCBI), with its deep focus on genomics and biological data banks, and the Lister Hill National Center for Biomedical Communications (LHC), a leader in clinical information analytics and standards.
Delivering Reliable, High Quality Biomedical and Health Information Services
NLM continues to expand the quantity and range of high quality information readily available to scientists, health professionals, and the general public. Advances in FY 2017included:
- indexing of approximately 1.1 million new journal articles for PubMed, NLM’s most heavily used database, which contains records for 27 million articles in biomedical and life sciences journals;
- growth in PMC, the digital archive of full-text biomedical literature, which now includes more than 4.5 million research articles, including those produced by researchers funded by NIH, 10 other Federal government agencies, and private research funders;
- expansion of gov, the world’s largest clinical trials registry, which now includes more than 256,000 registered studies and summary results for nearly 29,000 studies, including many not elsewhere published;
- enhancement of Genetics Home Reference (GHR), which provides consumer-level information on more than 2600 genetic conditions and genes to an average of more than 1.6 million visitors per month;
- twenty-four percent growth in dbGaP, which connects individual-level genomic data with individual-level clinical information and now contains more than 900 studies involving data from more than 1.5 million people;
- continued growth of PubChem, an archive of chemical and biological data on small molecules; PubChem contains information on more than 92 million unique chemical structures and more than 1.2 million bioassays;
- expansion of the RefSeq database of integrated, non-redundant, well-annotated reference sequences, which are essential to identifying and documenting genetic variations that affect human health, to over 140 million records, a 31 percent increase in FY 2017, including more than 95 million protein records from over 72,000 organisms;
- expansion of the AccessGUDID database by 50 percent to include FDA unique identifiers and registration information for nearly 1.5 million medical devices, supporting improvements in care and patient safety; usage more than doubled since its introduction in FY 2016, with nearly 15 million application programming interface (API) calls from computer systems accessing the resource.
- More than 76 million API calls to MedlinePlus Connect from health IT systems and EHRs, requesting patient-specific delivery of consumer health information from MedlinePlus
NLM also continues to expand access to its rare and unique historical collections through digitization partnerships with outside organizations. In FY 2017, more than 1,800 printed historical books were digitized and added to NLM’s Digital Collections, a free online archive of biomedical books and videos.
NLM enhances its computing technology infrastructure by taking advantage of advanced computing techniques, such as cloud storage, that offer greater agility, better resource utilization, and potentially lower cost. In FY 2017, NLM launched cloud implementations of PubMed Labs, BLAST for sequence searches, and the Common Data Element repository.
Program Portrait: ClinicalTrials.gov
ClinicalTrials.gov is the world’s largest, publicly accessible database for exploring clinical research studies conducted in the United States and abroad. The database lists information from more than 250,000 publicly and privately supported clinical studies from 201 countries, including summary results information for nearly 29,000 of these studies. It provides patients, their family members, health care professionals, researchers, and the public with easy access to information on clinical studies on a wide range of diseases and conditions. Study sponsors or investigators submit information to ClinicalTrials.gov when a study begins, and update the information throughout the study lifecycle. Listing of a study does not imply endorsement by the NIH or the Federal Government.
ClinicalTrials.gov promotes research accountability by increasing transparency across the clinical research enterprise. Broad dissemination of ongoing and completed research, including summary results, helps meet the ethical commitment to research participants who have no assurance of personal benefit, but who expect their participation to help others. Additional impacts include an increase in rigor and reproducibility, as well as the ability to improve research designs by learning from prior research. The database also enables NIH to provide optimal stewardship of the clinical research it funds by providing context for each study within the overall research landscape, and by facilitating the tracking of key metrics related to research output.
The scope and content of information required to be submitted to ClinicalTrials.gov was expanded in January 2017 under U.S. regulations and a new NIH Policy. Specifically, the Final Rule for Clinical Trials Registration and Results Information Submission (42 CFR Part 11) clarifies and expands the registration and results information submission requirements of the Food and Drug Administration Amendments Act of 2007. The NIH Policy on Dissemination of NIH-Funded Clinical Trial Information establishes the expectation of registration and results submission to ClinicalTrials.gov for all NIH-funded clinical trials. Growing awareness of these requirements and other international policies has led to a 10 percent increase in the rate of study registration and greater than 50 percent increase in rate of results submissions. To help ensure that this rich resource of information is accessible and useful to the public, NLM worked with 18F (a digital services consultancy in the General Services Administration) to enhance the usability of ClinicalTrials.gov by making it easier for users to search and retrieve studies of interest.
Promoting Public Awareness and Access to Information
When natural disasters strike, NLM has programs in place to provide access to health information throughout the country. This includes launching the Emergency Access Initiative that ensures free access to the published literature, and developing websites that pull together relevant information specific to each occurrence. In FY 2017, NLM provided information in support of hurricane disaster response in the southeastern U.S. and the Caribbean. Disaster response efforts included National Network of Libraries of Medicine (NNLM) participation at local levels, working to ensure that patients, families, and the general public have access to information that assists with addressing health needs where they are – in evacuation shelters, neighborhoods, library branches, and community centers.
NLM offers direct-to-consumer information resources in lay language. MedlinePlus includes information about disease, conditions, and wellness issues. In FY 2017, the number of health topics covered reached 1000. MedlinePlus information is also available through MedlinePlus Connect, which works with EHR systems to bring the information to patients and health care providers at the point of need in healthcare systems. Websites focused on consumer health information related to the environment, drug information, genetics, and specific populations are also offered.
NLM uses multiple channels to reach the public, including direct contact, development of consumer-friendly websites, and human networks that reach out to communities. NLM also uses novel approaches through technology engagement, such as with hackathons that use NLM resources to address specific information problems. In FY 2017, NLM funded more than 200 outreach projects across the country to enhance awareness and access to health information, and to address health literacy issues. It also hosted seven hackathons.
NLM uses exhibitions, the media, and new technologies in its efforts to reach underserved populations and promote interest among young people in science, medicine, and technology. NLM continues to expand its successful traveling exhibitions program, which is a cost-efficient way to extend access to NLM resources around the world, including international locales for access by U.S. Armed Forces. In FY 2017, public-private partnerships enabled NLM traveling exhibitions to appear in 216 institutions in 161 towns and cities in 45 states and two other countries. Examples include: Physician Assistants: Collaboration and Care; Native Voices: Native Peoples’ Concepts of Health and Illness; Confronting Violence: Improving Women’s Lives; and Fire and Freedom: Food and Enslavement in Early America.
As a collaborative venture with NIH Institutes and Centers (ICs) and other partners, NLM produces the NIH MedlinePlus magazine, and its Spanish counterpart, NIH Salud. The magazine, which is also available online in Spanish and English, is distributed to 70,000 individual and bulk subscribers, as well as to doctors’ offices, health science libraries, Congress, the media, federally supported community health centers, select hospital emergency and waiting rooms, and other locations nationwide where the public receives health services. In FY 2017, NLM and NIH partnered with the National Hispanic Medical Association, the Alzheimer’s Association, the American Diabetes Association, the Peripheral Arterial Disease Coalition, Research!America, and the Lymphatic Education and Research Network, among others, to extend the distribution of the magazine to the audiences they serve.
Program Portrait: National Network of Libraries of Medicine (NNLM)
The 6,500 member institutions of the NNLM are valued partners in ensuring that health information, including from NLM services, is available to scientists, health professionals, and the public. NNLM is coordinated by eight Regional Medical Libraries (RMLs) and is comprised of academic health sciences libraries, hospital libraries, public libraries, and community-based organizations.
The RMLs are supported through cooperative agreements and are located at health sciences libraries at the University of Massachusetts; the University of Pittsburgh; the University of Maryland; the University of Iowa; the University of Utah; the University of North Texas; the University of California, Los Angeles; and the University of Washington. Under the cooperative agreements, the NNLM is governed by a National Network Steering Committee (NNSC), which is working together to ensure coordination among regions for services that can be delivered nationwide by one library to all regions. The NNLM plays a pivotal role in outreach to NLM’s diverse audiences by exhibiting and demonstrating NLM's products and services at national, regional, and state health professional and consumer oriented meetings; coordinating efforts to improve access to electronic publications for the public health workforce; improving awareness and access to high quality health information for the general public; addressing health literacy and health disparities. In addition, the NNLM is partnering with the NIH All of Us Research Program to support community engagement efforts by public libraries across the United States and raise awareness about the program. The NNLM RD3: Resources for Data-Driven Discovery site was developed to support data science as a resource for information professionals to learn about library roles in data science, fundamentals of domain sciences, and emerging trends in supporting biomedical research. The NNLM established a partnership with the Public Library Association, Promoting Healthy Communities, to increase the capacity of public libraries to provide health information to patrons and help them learn about emerging health information needs in public libraries.
The NNLM has an excellent track record of providing access to health information in disasters and emergencies and will continue to serve as the backbone of NLM's strategy to promote more effective use of libraries and librarians in local, state, and national disaster preparedness and response efforts. NNLM also plays an important role in NLM efforts to increase the capacity of research libraries and librarians to support data science and improve institutional capacity in biomedical big data management and analysis.
Developing Advanced Information Systems, Standards, and Research Tools
NLM’s advanced information services have long benefitted from its intramural research and development (R&D) programs and from its efforts in promoting and supporting health data standards. Collectively, these efforts have led to major advances in the ways high volume information and data are collected, structured, standardized, mined, and delivered. The Library conducts advanced R&D on different aspects of biomedical informatics through LHC and NCBI.
Lister Hill Center for Biomedical Communications (LHC)
Established by joint resolution of Congress in 1968, LHC conducts and supports R&D in such areas as the development and dissemination of health data standards; the capture, processing, dissemination, and use of high quality imaging data; medical language processing; high-speed access to biomedical information; advanced technology for emergency and disaster management; and analysis of large databases of clinical and administrative data to determine their usefulness in predicting patient outcomes and in validating findings from relatively small prospective clinical research studies.
In the area of natural language processing, LHC conducts research involving language resources and innovative algorithms and develops tools to help advance the fields of natural language understanding and biomedical text mining and apply them to indexing and information retrieval. Projects include the UMLS (Unified Medical Language System), Medical Text Indexer (MTI), SemRep (Semantic Knowledge Representation Project), MetaMap, and most recently, MetaMap Lite. MetaMap Lite, released in FY 2017, is a fast, customizable, and easy to use tool for identifying medical symptoms, findings, risk factors, treatments, and diagnoses in free text narrative.
Leveraging extensive machine learning experience and field-based projects in processing clinical images from parasites to lungs, LHC is now advancing the analytical tools that are being applied in image analysis research. In FY 2017, LHC began applying a new machine learning technique, known as deep learning, or deep neural networks because they imitate the networks of neurons in biology. Deep learning improved performance in two ongoing projects: automated classification of chest X-rays in the field to detect patients with active tuberculosis, and automated detection and counting of parasitic cells in NLM’s MalariaScreener smartphone application for malaria detection in the field. It is also being applied in two new image analysis projects. In collaboration with the National Eye Institute (NEI), LHC scientists are studying a set of 51,700 retinal photographs to identify and measure the size of the lesions associated with diagnosing and managing retinal diabetes and age-related macular degeneration; these techniques make it easier to assess the effect of treatments in clinical care and research drug trials. In addition, deep learning techniques are being applied with a dataset of de-identified skin photographs to help classify skin diseases.
The National Center for Biotechnology Information (NCBI)
Established by law in 1988 (P.L.100-607), NCBI conducts R&D on the representation, integration, and retrieval of molecular biology data and biomedical literature, in addition to providing an integrated genomic information resource consisting of more than 40 databases for biomedical researchers at NIH and around the world. These databases range from data on human genetic variation and viral pathogens to information on genetic tests. NCBI’s development of large-scale data integration techniques with advanced information systems is key to its expanding ability to support the accelerated pace of research made possible by new technologies, such as next-generation DNA sequencing, microarrays, and small molecule screening. GenBank at NCBI, in collaboration with partners in the United Kingdom and Japan, is the world’s largest annotated collection of publicly available DNA sequences. GenBank contains 201 million sequences from more than 400,000 different species. NCBI’s web services for access to these data provide the information and analytic tools for researchers to accelerate the rate of genomic discovery and facilitate the translation of basic science advances into new diagnostics and treatments.
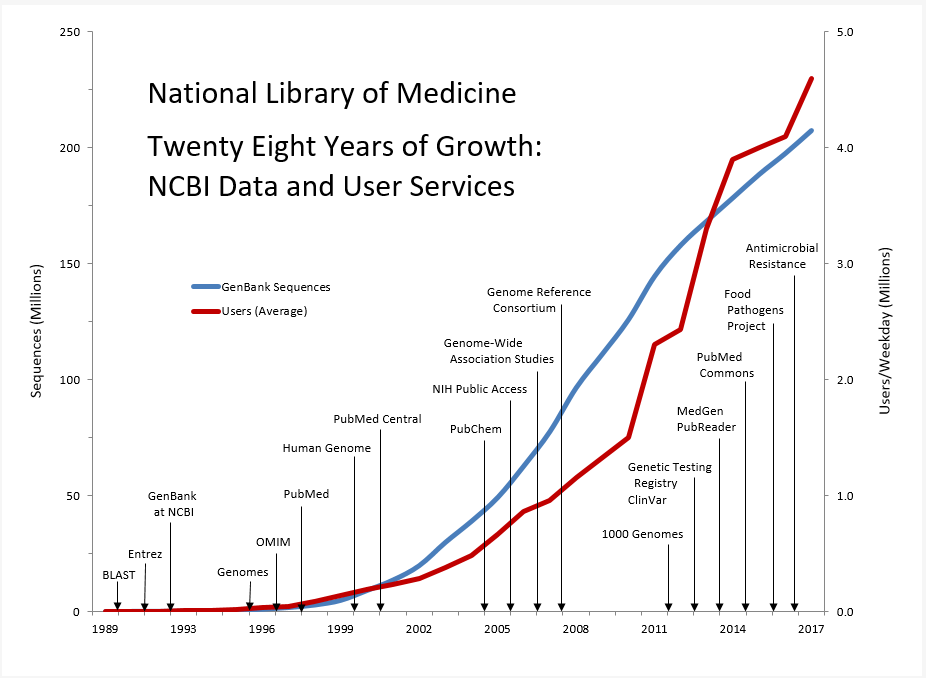
Data for Twenty Eight Years of Growth: NCBI Data and User Services
| Year | Product Release | GenBank Sequences | Users (Average) |
|---|---|---|---|
| 1989 | 30,010 | 0 | |
| 1990 | BLAST | 40,295 | 0 |
| 1991 | Entrez | 58,952 | 0 |
| 1992 | GenBank at NCBI | 87,846 | 0 |
| 1993 | 143,492 | 4,500 | |
| 1994 | 215,273 | 9,000 | |
| 1995 | Genomes | 555,694 | 19,000 |
| 1996 | OMIM | 1,021,211 | 36,000 |
| 1997 | PubMed | 1,765,847 | 39,268 |
| 1998 | 2,837,897 | 84,256 | |
| 1999 | Human Genome | 4,864,570 | 138,139 |
| 2000 | PubMed Central | 9,102,634 | 190,485 |
| 2001 | 13,602,262 | 233,977 | |
| 2002 | 19,808,101 | 283,590 | |
| 2003 | 29,819,397 | 380,104 | |
| 2004 | PubChem | 38,941,263 | 485,029 |
| 2005 | NIH Public Access | 49,152,445 | 666,917 |
| 2006 | Genome-Wide Association Studies | 62,765,195 | 864,586 |
| 2007 | Genome Reference Consortium | 77,632,813 | 958,584 |
| 2008 | 96,400,790 | 1,152,700 | |
| 2009 | 110,946,879 | 1,328,143 | |
| 2010 | 1000 Genomes | 125,764,384 | 1,501,335 |
| 2011 | 144,458,648 | 2,300,000 | |
| 2012 | Genetic Testing Registry and ClinVar | 157,889,737 | 2,430,000 |
| 2013 | MedGen and PubReader | 168,335,396 | 3,300,000 |
| 2014 | Pubmed Commons | 178,322,253 | 3,900,000 |
| 2015 | Food Pathogens Project | 188,372,017 | 4,000,000 |
| 2016 | Antimicrobial Resistance | 197,390,691 | 4,100,000 |
| 2017 | 207,265,929.5 | 4,600,000 |
As part of the National Action Plan for Combating Antibiotic-Resistant Bacteria (CARB), NCBI collaborates with FDA, CDC, USDA, and other groups to maintain a database of whole genome sequencing (WGS) data for antibiotic-resistant bacteria, along with tools to facilitate analyses of such data. The database provides an important resource for surveillance and research into the mechanisms underlying the emergence of antibacterial resistance. This program builds upon a successful collaborative project among these same agencies to use WGS to more quickly and accurately identify and investigate outbreaks of disease caused by foodborne bacterial pathogens such as listeria and salmonella.
The computational biology research that is rooted in complex analyses of richly annotated genomics data resources has yielded important discoveries and health advances. In FY 2017, NCBI researchers studying the range of gene-altering mutagens involved in cancer developed a new analytic method, called BeWith, in which gene modules with specific mutational patterns can be identified within large sets of patient data. In addition, the MutaGene resource was designed to help researchers and clinicians understand the mutagenic factors that change DNA and contribute to the development of tumors. In ongoing research on gene editing, scientists are using genome data resources and analysis algorithms to identify new CRISPR-Cas variants. In support of drug discovery, NCBI scientists developed a software tool called AptaTRACE for identifying aptamer molecules, which are short segments of DNA or RNA that are capable of binding with high precision and specificity to targets of interest, based on features of their sequence and structure. This makes them highly useful in therapeutics, diagnostics, and drug development. AptaTRACE analyzes the molecular sequence and 3-D structure of the aptamers and the various targets to which they bind in order to determine and understand the common features (or “motifs”) that are involved in binding. Such information allows researchers to understand better why some molecules bind and others do not. This research is an excellent example of how the benefits of big data critically depend upon the existence of algorithms that are capable of transforming such data into information.
Health data standards
NLM has been a major force in health data standards for more than 30 years. In close collaboration with the Office of the National Coordinator for Health Information Technology within HHS and with support from CMS, the Veterans Health Administration, and FDA, NLM develops, funds, and disseminates the clinical terminologies designated as U.S. standards for meaningful use of EHRs and health information exchange. This includes three standard vocabularies— LOINC (for identifying tests and measurements), RxNorm (for identifying drugs), and SNOMED CT (for identifying problems, organisms, and many other special items). The goal is to ensure that EHR data created in one system can be transmitted, interpreted, and aggregated appropriately in other systems to support health care, public health, and research. NLM produces a range of tools that help EHR developers and users to implement these standards and makes them available in multiple formats, including via application programming interfaces (APIs). Importantly, NLM’s financial support also allows key standards to be used free-of-charge in U.S. health care, public health, biomedical research, and product development.
NLM’s Unified Medical Language System (UMLS) resources connect standard clinical terminologies to billing codes and more than 120 other important biomedical vocabularies, such as those used in information retrieval and gene annotation. By linking many different names for the same concepts and by providing associated natural language processing tools, UMLS resources help computer programs to interpret biomedical text and health data correctly in NIH-funded research, in commercial product development, and in many electronic information services, including those produced by NLM.
In FY 2017, a new specification for defining the mappings from laboratory instruments’ transmission codes to appropriate LOINC codes for identifying medical laboratory observations was produced via collaboration among NLM, CDC, FDA, Regenstrief Institute, and the In-Vitro Diagnostics Industry Connectivity Consortium (IICC). This work will greatly facilitate the pooling of laboratory results from different sources about the same patient, across multiple laboratory sources and care venues and will underpin many NIH initiatives, including All of Us and the Cancer Moonshot.
RxNorm is a widely used drug terminology developed by NLM and used for electronic prescription and exchange of drug information. NLM has developed a graphical user interface (RxNav) and APIs to facilitate access by researchers, industry, and the public. In FY 2017, these drug APIs received 800 million queries. Recent developments include support for analytics to support longitudinal analysis of prescription datasets from claims and EHR systems.
Extramural Programs
NLM funds extramural research, resource, and workforce development programs that provide important foundations for the field of biomedical informatics and data science, which brings the methods and concepts of computational, informational, quantitative, social/behavioral and engineering sciences to bear on problems related to basic biomedical/behavioral research, health care, public health, and consumer use of health-related information. In addition to grants for basic and applied research, predoctoral and postdoctoral training, and career development, NLM sponsors several unique resource grant programs that support biomedical knowledge resource development. To accomplish its extramural goals, NLM offers grants in four general categories: research project grants and supplements; training, fellowship, and career support; information resource awards; and small business grants. NLM will also continue to provide management oversight for a selection of grants for NIH pioneer and early innovation awards and data science research training and digital curation awards, funded by the NIH Common Fund.
Innovation in data science will require significant expansion of the extramural program of the NLM over time. Research is needed to uncover the places and problems where data science challenges remain, and to create principled, extensible solutions to these challenges. Additional investment is needed to extract generalizable learnings from the analytical results and visualization tools devised through existing domain-specific and problem inspired research, ensuring that such advances are sufficiently robust to take on problems not yet envisioned or encountered.
Informatics Workforce and Resources for Biomedicine and Health
Many of today’s informatics researchers and health information technology leaders are graduates of NLM-funded university-based training programs in biomedical informatics. As of July 2017, NLM supported research training in biomedical informatics and data science at 16 active university-based programs, training more than 170 individuals each year, including 14 trainees emphasizing environmental exposures funded by NIEHS. In addition, 14 grant supplements were awarded to NLM’s university-based training programs for curriculum and faculty development in data science. NLM also supports individual predoctoral fellowships via the National Research Service Awards (NRSA) program; three new awards were made in this program in FY 2017. Two career transition programs are offered to NLM’s trainees and others ready to launch their informatics research careers. Taken together, NLM’s commitment to training and career transition in FY 2017 represented nearly 33 percent of NLM’s extramural grants budget. In FY 2018, NLM will continue to support research training and career transition through institutional training programs and grants to individuals.
NLM Administrative Supplements for Informationist Services provide supplemental funds to existing NIH research grantees who want to add an information specialist to their research team. Three new informationist supplement awards were made in FY 2017, with librarians working on research teams looking at environmental exposure to manganese in rural areas, measurement tools for community-based participatory research, and engagement in clinical translational research.
The third unique program, NLM Information Resources to Reduce Health Disparities, has a unique focus on development of information resources; four three-year awards supporting development of information resources tailored to needs of Alaska Native, Navajo, and other underserved populations continued in FY 2017. Resources like these support long-term goals of the All of Us Research Program to engage a large cohort of active participants. In FY 2017, NLM awarded five three-year information resource projects.
Biomedical Informatics Research
NLM’s research project grants (RPGs) support pioneering research and development to advance knowledge in biomedical informatics and data science. Complementing and building upon informatics-related initiatives at other ICs, NLM research grant programs continue to support innovation in both basic and applied research ranging from small proof-of-concept projects to large, multi-site research collaborations. NLM has also launched a new grant initiative using data science concepts and approaches to solve problems consumers face in accessing, storing, using, and understanding their own health data. This work will produce tools that make data science findings more understandable to patients. NLM funds investigator-initiated projects, as well as projects from focused funding announcements that target areas important to NLM’s mission.
Program Portrait: Research in Biomedical Informatics and Data Science
For more than 30 years, NLM’s Extramural Programs Division has been a principal source of NIH support for research in basic and applied biomedical informatics and data science. NLM’s portfolio of funded research has spanned artificial intelligence, computational biology, clinical decision support, public health surveillance, and visualization and discovery mining in digital data sets. The scope of NLM’s research interests is wide, encompassing informatics and data science research areas of high importance to NIH and society at large, and for audiences ranging from clinicians and scientists to consumers and patients.
In recent years, NLM has funded research project grants that address important data science issues, including:
- accounting for bias, missing data, and other inconsistencies when using data from EHRs in research;
- enhancing the value of existing data from intensive care units for real-time decision making by understanding the timing of gradual state changes and validating causation assertions made using uncertain data;
- testing methods used to extract information from published articles about relationships among drugs, genes, and phenotypes to see if they are also effective on text in EHRs;
- re-engineering precision therapeutics through N-of-1 trials;
- developing tools to help researchers understand the heterogeneity of single cancer cells in a tumor by allowing multiple genomic assays of a single cell, to assist in identifying effective cancer therapies;
- estimating risk of foodborne illness using public health and social media sources;
- designing and testing tools to simplify text for patients and consumers; and
- biomedical computing and informatics strategies for precision medicine.
NLM plans to continue to fund research on a wide range of biomedical informatics and data science questions. In addition, NLM aims to expand its research into analytic approaches and visualization tools that help patients, consumers, and clinicians participate actively in precision medicine research.
In FY 2017, NLM issued 24 new RPGs, including six exploratory/developmental awards. Awards reflect current and expanding investments in data science, as well as investment in data science applications for patients. Several of the new awards address data analytic topics, including integrating multiple data types; mechanistic machine learning; image classification; and EHR data analysis to improve comparative effectiveness research. Four new awards in translational bioinformatics focus on molecular interaction, gene regulation, genetic variants and neuroanatomical shape, and alterations of gene expression patterns. Among the newly funded research awards are also three in a new program that focuses on patient engagement in managing personal health information, including application of data science methods for improving patient and caregiver engagement and personal health records for individuals with multiple chronic conditions and for youth leaving from foster care. In support of the NIH Next Generation Researcher initiative, NLM awarded new research project support to two early stage investigators and two early established investigators.
NLM sets aside funds to support small business innovation and research and technology transfer (SBIR/STTR). In FY 2017, NLM met its required set-aside by funding three new SBIR/STTR awards, in addition to one continuing award; NLM’s allocation of funds for SBIR/STTR was more than $1 million. The new projects center on improvements to management and use of EHR data, application of data science and technology to phenotypic screening for drug discovery, and a mobile communication system for improved donor organ management and organ transplant outcomes.
Research Management and Support (RMS)
RMS activities provide administrative, budgetary, logistical, and scientific support for basic library services, intramural research programs, and the review, award, and monitoring of research grants and training awards. RMS functions also include strategic planning, coordination, and evaluation of NLM’s programs, regulatory compliance, policy development, and international coordination and liaison with other Federal agencies, Congress, and the public. These activities are conducted by the NLM Director and immediate staff, as well as NLM personnel from the Office of Extramural Programs, the Office of Administrative Management, the Office of Health Information Programs Development, and the Office of Communications and Public Liaison.
Detail of Full-Time Equivalent Employment (FTEs)
| OFFICE/DIVISION | FY 2017 Final | FY 2018 Annualized CR | FY 2019 President's Budget | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Civilian | Military | Total | Civilian | Military | Total | Civilian | Military | Total | |
| Division of Extramural Programs | |||||||||
| Direct: | 19 | - | 19 | 19 | - | 19 | 19 | - | 19 |
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Total: | 19 | - | 19 | 19 | - | 19 | 19 | - | 19 |
| Division of Library Operations | |||||||||
| Direct: | 258 | - | 258 | 258 | - | 258 | 258 | - | 258 |
| Division of Library Operations | |||||||||
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Division of Library Operations | |||||||||
| Total: | 258 | - | 258 | 258 | - | 258 | 258 | - | 258 |
| Division of Specialized Information Services | |||||||||
| Direct: | 35 | - | 35 | 35 | - | 35 | 35 | - | 35 |
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Total: | 35 | - | 35 | 35 | - | 35 | 35 | - | 35 |
| Lister Hill National Center for Biomedical Communications | |||||||||
| Direct: | 54 | - | 54 | 54 | - | 54 | 54 | - | 54 |
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Total: | 54 | - | 54 | 54 | - | 54 | 54 | - | 54 |
| National Center for Biotechnology Information | |||||||||
| Direct: | 290 | 1 | 291 | 297 | 1 | 298 | 297 | 1 | 298 |
| Reimbursable: | 4 | - | 4 | 4 | - | 4 | 4 | - | 4 |
| Total: | 294 | 1 | 295 | 301 | 1 | 302 | 301 | 1 | 302 |
| Office of the Director/Administration | |||||||||
| Direct: | 56 | - | 56 | 56 | - | 56 | 56 | - | 56 |
| Reimbursable: | 17 | - | 17 | 17 | - | 17 | 17 | - | 17 |
| Total: | 73 | - | 73 | 73 | - | 73 | 73 | - | 73 |
| Total | 733 | 1 | 734 | 740 | 1 | 741 | 740 | 1 | 741 |
| Includes FTEs whose payroll obligations are supported by the NIH Common Fund. | |||||||||
| FTEs supported by funds from Cooperative Research and Development Agreements. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FISCAL YEAR | Average GS Grade | ||||||||
| 2015 | 11.5 | ||||||||
| 2016 | 11.5 | ||||||||
| 2017 | 11.9 | ||||||||
| 2018 | 11.9 | ||||||||
| 2019 | 11.9 | ||||||||
Detail of Positions 1
| GRADE | FY 2017 Final | FY 2018 Annualized CR | FY 2019 President's Budget |
|---|---|---|---|
| Total, ES Positions | 5 | 5 | 5 |
| Total, ES Salary | 894,533 | 910,635 | 925,296 |
| GM/GS-15 | 25 | 25 | 25 |
| GM/GS-14 | 49 | 49 | 49 |
| GM/GS-13 | 142 | 149 | 149 |
| GS-12 | 113 | 113 | 113 |
| GS-11 | 34 | 34 | 34 |
| GS-10 | 0 | 0 | 0 |
| GS-9 | 10 | 10 | 10 |
| GS-8 | 38 | 38 | 38 |
| GS-7 | 11 | 11 | 11 |
| GS-6 | 2 | 2 | 2 |
| GS-5 | 0 | 0 | 0 |
| GS-4 | 2 | 2 | 2 |
| GS-3 | 3 | 3 | 3 |
| GS-2 | 2 | 2 | 2 |
| GS-1 | 1 | 1 | 1 |
| Subtotal | 432 | 439 | 439 |
| Grades established by Act of July 1, 1944 (42 U.S.C. 207) | 0 | 0 | 0 |
| Assistant Surgeon General | 0 | 0 | 0 |
| Director Grade | 0 | 0 | 0 |
| Senior Grade | 1 | 1 | 1 |
| Full Grade | 0 | 0 | 0 |
| Senior Assistant Grade | 0 | 0 | 0 |
| Assistant Grade | 0 | 0 | 0 |
| Subtotal | 1 | 1 | 1 |
| Ungraded | 283 | 283 | 283 |
| Total permanent positions | 424 | 431 | 431 |
| Total positions, end of year | 732 | 739 | 739 |
| Total full-time equivalent (FTE) employment, end of year | 734 | 741 | 741 |
| Average ES salary | 178,907 | 182,127 | 185,059 |
| Average GM/GS grade | 11.9 | 11.9 | 11.9 |
| Average GM/GS salary | 102,841 | 104,692 | 106,378 |
1 Includes FTEs whose payroll obligations are supported by the NIH Common Fund.
Last Reviewed: August 20, 2024

